Assessing the integrity of RNA samples with the Bioanalyzer can be time-consuming and expensive since each run takes an hour and only 12 RNA can samples can be run.
To make RNA QC more convenient and affordable we will be running one or more batches of RNA samples weekly on the high-throughput LabChip GX.
For UC Davis labs the cost per sample will be $5 (with a minimum of $10).
In order to generate usable data:
- Provide sample names with an implicit order. The plots of the traces will identify the well position not sample names.
- We will not adjust sample concentration or volumes. It is your responsibility to meet the sample requirements.
- Submit total RNA samples in a well-sealed 96-well plate or in strip tubes (please see this page for examples) and a filled out QC submission form.
- For large sample numbers make sure that the plates are clearly labeled. Please use the QC submission form simply as a cover sheet in this case, listing the plate names and the number of samples for each. Samples should be filled into plates in column order from A1 to H1, then A2 to H2, etc. …
- Each RNA sample needs to have a volume 2 ul to 6 ul and contain 30 ng to 250 ng total RNA (this is the amount, not the concentration).
- Glycogen can interfere with the RNA QC and should be avoided (it can interfere with both spectrometry as well as capillary electrophoresis on the LabChip and the Bioanalyzer).
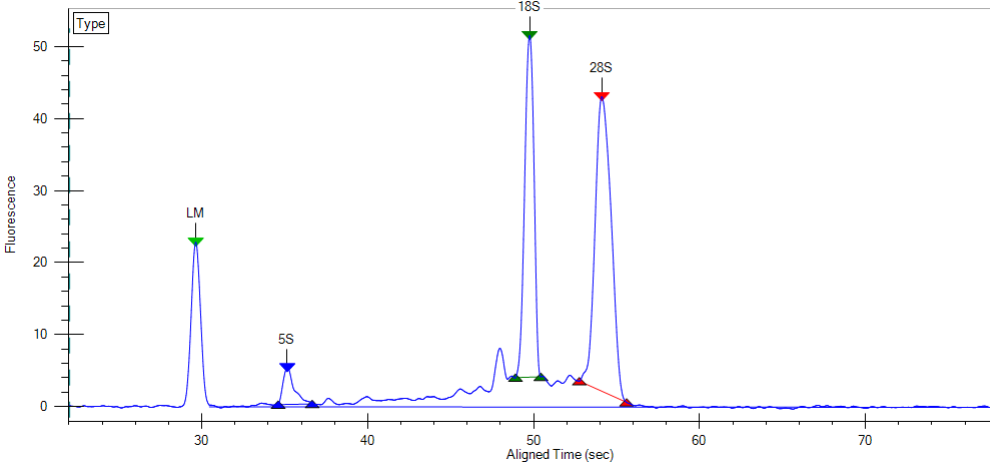
The LabChip GX will generate traces like the one below and RNA Quality Scores which are similar to the RIN scores provided by the Agilent Bioanalyzer and can be used interchangeably.
Please note that the capillary electrophoresis will be of lower resolution compared to the Bioanalyzer, but “good enough”. There is also no dedicated quality score algorithm for plant samples on the LabChip. The scores it produces are still realistic.
← FAQ