Dear DNA Technologies Core users,
This newsletter brings you announcements of: a new instrument for high quality DNA extraction and purification; two relevant CLC sequence analysis seminars tomorrow; an update on our April workshop on PacBio Library Prep and Data Analysis; and a Illumina HiSeq sequencing cost comparison.
UPDATES/NEWS
BorealGenomics Aurora DNA isolation/purification
We have added an BorealGenomics Aurora instrument to our core for high quality DNA extraction and purification:
The Aurora system enables the extraction DNA of high molecular weight DNA (up to megabase fragments) and the purification from difficult to remove contaminants. Please find more information here: http://www.borealgenomics.com/aurora/applications/ and example protocols here:
http://www.borealgenomics.com/customer-service/
Please email us if you would like to test the system.
Tomorrow: Qiagen presents two seminars on RNA-seq, gene annotation, and PAcBio data analysis with CLC-bio.
Session 1: Meyer Hall, Thursday, February 26th; 10 AM – 11.30 AM; Weir Conference Room, #2154;
Topics: CLC Genomic Workbench, Genome Annotation using Transcriptomic Reads, Microbial Genome Assembly and Utilization of Long Reads, Microbial Genome Assembly and Utilization of Long Reads
Session 2: Genome Center, Thursday, February 26th 2 PM – 3.30 PM, Room #4202
Topics: Overview of Tools and Applications, New Features in Latest Software Release, RNA-Seq using de novo assembled transcriptomes, Utilization of PacBio Reads for De novo Assembly
UC Davis Genome Center Workshop on PacBio Sequencing and Data Analysis (Wet lab and Bioinformatics)
PacBio sequencing is the method of choice for de novo assembly of high quality genome sequences (see these AGBT presentations), the sequencing of long amplicons and complex repetitive regions, the analysis of gene isoforms, and structural variant detection. With support from Pacific Biosciences, the DNA Technologies and Bioinformatics Cores are offering a sequencing library preparation and data analysis workshop for this revolutionary technology – to our knowledge, the first such workshop worldwide. The library preparation portion of the workshop will take place on April 14th and 15th, 2015, in the DNA Technologies Core Facility in the Genome Center (UC Davis). Hands-on data analysis using the PacBio SMRT-Portal will be explored on April 16th. The courses will begin at 8 am and end at 6 pm.
Please find the workshop registration page HERE
The library prep portion will cover the complete workflow starting from isolated DNA: sample QC, DNA shearing, library prep steps, library size selection and library QC. Libraries will be generated from genomic DNA samples. The participants will be able to work with their own sample and have the chance to generate a PacBio library, ready to sequence, for their research projects. Further, participants will be introduced to the details of the SMRT sequencing technology and other applications of the technology will be discussed. Participants will gain experience analyzing their PacBio sequencing reads using PacBio’s SMRT-Portal software, via virtual machines running on Amazon Web Services (AWS). We will use both public example data, and reads obtained from the DNA Technologies Core’s PacBio RS II, from libraries created during the workshop. Exercises will focus on running AWS virtual machines, data transfer and import, and assembly and variant finding protocols. Participants should be able to walk away from the workshop capable of analyzing their next PacBio data set independently.
The registration fee for the three-day workshop is $500, which will include all workshop materials and lunches. The workshop will be limited to 10 participants. Attendees do not need to be affiliated with the UC Davis campus.
To create the best possible libraries from your samples we highly suggest that you ship the samples at you earliest convenience to the DNA Technologies core for a QC of the DNA – we will run the sample on on a pulsed field electrophoresis gel. We will then inform you, if your sample will be suitable for PacBio sequencing. The DNA quality is essential for single molecule sequencing. We will provide control DNA samples in case you do not want to work with your own samples. For information on the sample requirements please see our page here: http://dnatech.genomecenter.ucdavis.edu/pacific-biosciences-rs/ . The shipping instructions are available here: http://dnatech.genomecenter.ucdavis.edu/sample-submission-scheduling/
Registrants are entitled to a 75% refund of the total registration price if cancelling no later than 2 calendar weeks before the start of a workshop. Within 2 weeks of workshop dates, no refund will be given. In order to cancel, you must send a written request to dnatech@ucdavis.edu within the allowed time. (Note that you may substitute another person from your organization at no charge, but please advise us as soon as possible so we can update our contact records.)
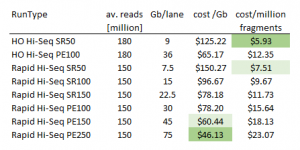
Cost comparison of HiSeq run types
Perhaps surprisingly the HiSeq Rapid Runs (paired end 150bp and paired end 250bp) provide the most Gb per dollar. Thus, rapid runs are not only of interest when longer reads are desired but also for applications that require highest possible coverage. HO indicates the default High-Output mode in the table below.