- Please make sure that you have PPMS account with the updated billing information (see the “Getting Started” page).
- Verify that your samples fulfill all the requirements.
- Please find the sample submission forms in our web-based CoreOmics portal : ucdavis.coreomics.com/dnatech/submissions/create
- You can copy-and-paste data from Excel or Google Sheets into the sample spreadsheets of the submission system.
- Select the type of project on the CoreOmics page here:
- Submission type choices are:
User-submitted libraries for Illumina or AVITI sequencing: Short-read sequencing user-submitted libraries — Pre-pooled or un-pooled.
DNA samples for short-read sequencing (Illumina & AVITI), RNA samples for short-read sequencing (Illumina & AVITI), 3′-Tag-Seq RNA-seq library prep and sequencing, PacBio sequencing, Nanopore PromethION sequencing, Single Cell (10X Genomics, ParseBio), QC & Quantification (Bioanalyzer, LabChip Gx, Tapestation, Qubit, etc.); Custom Processing (Custom & other services)
- Fill in the information electronically and print a copy to accompany your samples (landscape orientation, all on one page). Samples can only be processed if we receive a hard copy of your submission form together with them. The CoreOmics system will prompt you to print.
- We require Bioanalyzer traces (or similar) to work with customer submitted sequencing libraries as well as for total RNA samples. If no traces are submitted with the libraries/samples, we will carry out the Bioanalyzer QC for a fee. If you can supply traces, please upload them to the CoreOmics submission system and/or include them into the shipment in hard copy.
Please make sure that the samples conform to the sample or library requirements: see this page. - Submit your samples in 1.5 ml microcentrifuge tubes. The tubes have to be clearly labeled on the lid. We suggest using low-retention tubes (e.g. Eppendorf LoBind). Please protect these tubes in a sturdy box or in disposable 50 ml centrifuge tubes from damage from cold-packs. If using wet ice, double-pack your samples in two containers or sealed bags. Make sure that the lids can not loosen by adding padding on top of them. Do not seal the tubes with Parafilm.
- RNA samples for large-scale projects can be submitted in strip tubes with individually attached caps (RNAse-free). Please pack the strips into “racks” (e.g. empty pipet tip boxes) and make sure that they can’t move during transport.
 .
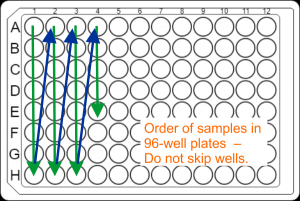
. - DNA samples for large-scale projects should be submitted in the strip tubes described above, but can also be submitted in well-sealed 96-well semi- or fully-skirted PCR plates (best Eppendorf twin.tec PCR plate 96 LoBind). Sample placement in plates: Samples have to be placed in the column orientation ( A1, B1, C1, D1, …. , A2, B2, C2, D2, ….) in contiguous wells. Do not skip wells, rows, or columns.
 To ensure that your samples arrive without leakage and cross-contamination, use one of the following types of seals: Cap the wells with matching 8-strip caps. These are usually ordered separately from the plates. Be sure that the caps seal tightly! Foil seals: Heat Seal (this is the preferred seal method. Please use a removable heat seal that allows the plate to be resealed such as “Thermo Scientific Easy Peel Heat Sealing Foil”) or adhesive aluminum foil seals like “AlumaSeal CS Film”. Protect the plates in a sturdy box with plenty of cushioning. Sample shipments of plates should be carried out on a surplus of frozen “blue ice” blocks or on dry ice, to insure that the samples remain frozen at all times through the shipment.
To ensure that your samples arrive without leakage and cross-contamination, use one of the following types of seals: Cap the wells with matching 8-strip caps. These are usually ordered separately from the plates. Be sure that the caps seal tightly! Foil seals: Heat Seal (this is the preferred seal method. Please use a removable heat seal that allows the plate to be resealed such as “Thermo Scientific Easy Peel Heat Sealing Foil”) or adhesive aluminum foil seals like “AlumaSeal CS Film”. Protect the plates in a sturdy box with plenty of cushioning. Sample shipments of plates should be carried out on a surplus of frozen “blue ice” blocks or on dry ice, to insure that the samples remain frozen at all times through the shipment. - Please use unique sample names! (e.g. three-letter codes plus numbers).
- For genomic DNA and other dsDNA shipments in tubes, cold packs (e.g. “blue ice”) are usually sufficient.
- RNA samples should be shipped on dry ice. Please only ship with courier services (use FedEx, UPS — do NOT use DHL). For longer transports we also had success with RNA samples shipped dry at room temperature (after LiCL/ethanol precipitation and ethanol washes; please note mark the position of the pellet on the tubes).
- If you are shipping DNA for PacBio libraries, please follow the PacBio guidelines for shipping and handling.
- Do not ship any samples before a weekend (i.e., on a Thursday for Friday arrival) or the day before a holiday: see this calendar of our University holidays .
- Please drop-off the samples in our lab (GBSF, #1410) or use the following shipping address and ship samples via courier services (FedEx, UPS, DHL) :
- We highly suggest using the “Priority Overnight Shipping” options for delivery in the morning, since these seem more reliable.
- NEVER use US Postal Mail to send samples, such shipments will get delayed severely while being moved at a snail’s pace on campus.
For international shipments: UC Davis EIN-number # 94-6036494
Please contact us if you have any questions about the required information. If you drop off your samples in person we can go over the submission form with you. It is essential that you fill in all the appropriate information to minimize the chance of error on these expensive and time-consuming experiments. If we are not in the lab when you come by, please deposit your samples in the white polystyrene cool box on the lab bench – do not deposit them in the under-counter freezer or remove any samples from this freezer.
Demultiplexing – It is important to ensure your sample IDs and barcodes are unique – we are now implementing additional labor charges for re-demultiplexing because of incorrect submission info, duplicate sample names, wrong barcodes, reverse complement sequence in error, etc.
Scheduling
The turnaround times can be difficult to predict far in advance, since it depends on the requested options and the queue length at time of sample drop-off. Thus we can usually give a better estimate when your sample is delivered. For MiSeq runs the turnaround time is typically five to eight days, while you should allow four to six weeks for the HiSeq sequencers. Libraries that are ready-to-go are usually sequenced within 3 weeks. Please see our HiSeq Calendar & Queue for the projected HiSeq sequencing schedules. We do encourage scheduling discussions at the outset of your project since, depending on the requirements of your run, we may need to order particular reagents. This is particularly true for non-standard ribo-depletion requests and some MiSeq runs as the 50 and 300 cycle kits are not kept in stock and need to be special ordered.

