The Pippin systems from SageScience allow for precise automated size selection of nucleic acid samples, sequencing libraries, and even protein samples. Our Core runs both the BluePippin and the Pippin HT. Whereas the former enables selection over the widest size ranges, the Pippin HT excels at high-throughput applications and also works with smaller sample amounts..
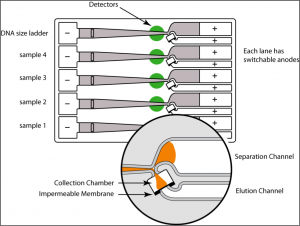
The BluePippin system uses dye-free, pre-cast gel cassettes to separate and extract desired DNA fragment size ranges from agarose. It enables size selection for fragment lengths between 90 bp to 50 kb using different gel cassettes. The BluePippin system can run pulsed-field gel electrophoresis programs and thus separate large DNA fragments. Depending on the type of gel cassette up to 4 or 5 samples/cassette may be run, with no possibility of cross contamination.
Samples in standard buffers (e.g., TE, EB) are compatible with the gel buffer, but preps in high-salt solutions (e.g., after excess concentration in a speed-vac) have to be avoided. The samples also have to be detergent-free. Do not load PCR reactions directly into Pippin instruments, since PCR buffers tend to contain detergents.
The BluePippin agarose cassettes available in the Core are:
0.75% for PacBio, external markers, S1 (< 20 kb), U1 (>30 kb)
1.5% internal markers, 250 bp – 1.5 kb
2% internal markers, 100 bp – 600 bp
3% for miRNA, internal markers, 90-250 bp
The BluePippin loading volume is up to 30 ul of DNA dissolved in a low salt buffer (like EB, TE, or most enzyme buffers, or water).
For example Iso-Seq cDNA samples should contain at least 2 ug to 5 ug DNA in a 30 ul volume.
Genomic DNA samples/PAcBio libraries should contain at least 2.5 ug to 5 ug DNA in a 30 ul volume.
Please also see the BluePippin cassette reference chart from SageScience.
Our assumption is that we lose about 50% of the DNA of the desired lengths during a BluePippin size selection, thus a considerable amount of input material is required. We recommend starting with 1 to 5 ug in a max volume of 30 ul (although as low as 500ng in 30 ul is possible in some cases).
The BluePippin instrument is available both as a Core Service and to campus users as shared equipment, after training.
The Pippin HT sample sizing instrument allows to precisely size-select DNA samples and sequencing libraries for up to twenty-four samples per single run.

The instrument belongs to our Shared Equipment – this means all UCD researchers can be trained to use this instrument.
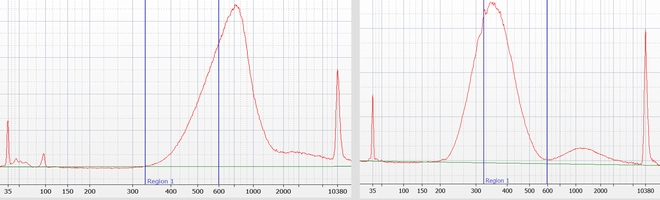
We are using the instrument for precise size-selection for all genomic DNA sequencing libraries prepared by our lab. This allows us to avoid generating redundant data for short insert library molecules (for which the reads would otherwise overlap); the precise size selection will further avoid lower quality data caused by library molecules that are too long (e.g. longer than 600 bp for the HiSeq 4000). For the two example customer libraries size selection for the optimal sizes (within the blue bars) removed about 50% of the library molecules that are outside of the optimal range.
The Pippin HT agarose cassettes available in the Core and the size ranges in which samples can be sized:
0.75% from 6 kb up to 40 kb high -pass
1.5% from 250 bp up to 1.5 kb
2% from 100 bp up to 600 bp
For the the 2% cartridge selection intervals can be as small as 20 bp.
The Pippin HT loading volume is up to 20 ul of DNA dissolved in a low salt buffer (like EB, TE, or most enzyme buffers, or water).
Samples in the typical Illumina library size range should contain at least 200 ng DNA in a 20 ul volume (detergent-free).
Please refer to the PippinHT manual for more details.