The DNA Technologies Core discontinued Illumina microarray/Infinium genotyping services in October 2023.
Please contact the UC Davis Veterinary Genetics Lab (VGL; https://vgl.ucdavis.edu/ ; phone: (530) 752-2211 ) for Illumina microarray services.
FOR REFERENCE ONLY:

 The Infinium platform from Illumina is an extremely high throughput SNP genotyping system that allows the identification and scoring of up to 2.5 million SNPs per single DNA sample. Unlike the Golden Gate assay (now discontinued) that uses universal primers to amplify SNP-reactive DNA fragments, the Infinium assay relies on direct hybridization of genomic targets to array-bound sequences. Single base extension is followed by fluorescent staining, signal amplification, scanning, and analysis using the Genome Studio software.
The Infinium platform from Illumina is an extremely high throughput SNP genotyping system that allows the identification and scoring of up to 2.5 million SNPs per single DNA sample. Unlike the Golden Gate assay (now discontinued) that uses universal primers to amplify SNP-reactive DNA fragments, the Infinium assay relies on direct hybridization of genomic targets to array-bound sequences. Single base extension is followed by fluorescent staining, signal amplification, scanning, and analysis using the Genome Studio software. 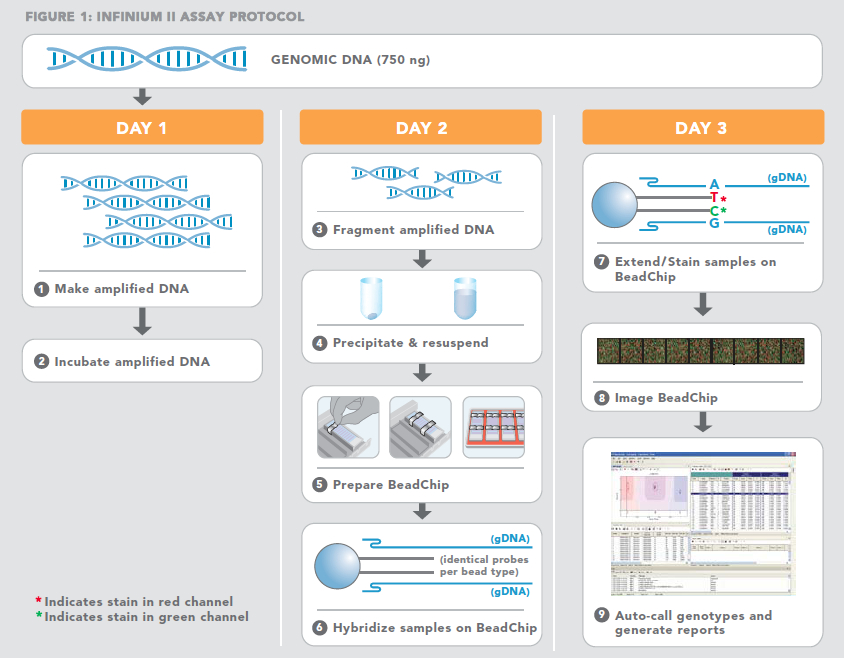 A schematic of the Infinium assay is shown below.
A schematic of the Infinium assay is shown below.
Due to the high number of SNPs that can be analyzed on Infinium chips, simplicity of sample preparation, and relative ease of data analysis, this platform has been widely exploited for dozens of major studies in human genetics. These include whole genome association studies, population genetic analyses, and copy number variation investigations, to name a few.
Numerous configurations of human sequence-based Infinium chips are available, designed to satisfy the different needs of particular studies. We can’t comment on the scientific rationale for selection of a particular chip type. However, we can mediate interactions with the Illumina technical support staff to design a research plan.
In addition to providing arrays facilitating human genetic studies, Illumina offers a number of products for analysis of agriculturally important species including maize, cows, and pigs. For a more comprehensive listing of available catalog genotyping options, check out Illumina’s web site. Illumina also will design and produce custom Infinium chips, called iSelect, which can query between 3,000 and 200,000 SNPs/sample. This provides customized genetic analysis at a level unavailable through other platforms or assays and has been exploited successfully by several of our users.
Beginning the Project
The first step is to determine the SNPs to be queried; after a SNP list is generated, it is submitted to Illumina. Typically a back-and-forth process between you and Illumina will ensue to select the best set of SNPs for the Infinium assay. This process is described in more detail in this tech note on assay design. Once the final SNP list is ready, it is emailed to us and we place the order with Illumina for your materials. Generally this process of design and ordering will involve an ongoing dialogue among Core personnel, the local Illumina rep, your lab, and Illumina tech support.
Sample Submission
For most Infinium assays 4 ul of 50 ng/ul DNA is the recommended input material. We request at least 10 ul of DNA at this concentration. Please provide clean and accurately quantified input genomic DNA. Some degradation is tolerated, but extensively degraded material may not perform well in the assay. Illumina recommends that all DNA be quantified using the PicoGreen fluorometric assay – our facility has the reagents and equipment necessary to carry out this Picogreen DNA quantification assay, please inquire if you’re interested in this option. In general we find that Nanodrop readings plus gel visualization of all samples, if feasible (if not, viewing a subset of the samples by gel electrophoresis is the next best thing), provides sufficient quality control determination for DNA to be used in the assay. Samples should be resuspended in water, 10 mM Tris 8.0, Qiagen EB (10 mM Tris 8.5), or TE. It is essential to maintain a stock of the same DNA samples in your lab if needed. By default we will dispose of excess DNA unless arrangements are made to pick up your remaining samples.
Samples should be submitted in a 96-well plate and accompanied by a sample submission template. The form must be filled to completion, please contact us if you have any questions about the required info. Depending on the chip layout, different templates are required. Please download the appropriate form for your project, as indicated below:
Infinium 24 x 1 Sample Submission Form
Infinium 12 x 1 Sample Submission Form
Infinium HD Super Quad Sample Submission Form
Infinium 8 x 1 LCG Sample Submission Form
Output and Analysis
Data produced from Infinium assays must be analyzed initially using Illumina’s Genome Studio software. There are various ways for our customers to access this software, please contact us for details. The software can analyze and export data in a variety of formats, from a matrix of genotype calls to a tabulation of the raw red and green intensities for every SNP in every sample. Careful experimental design regarding sample preparation and selection, as well as considerations of the downstream data analysis of the datasets produced, will require thought and expert scientific input beyond the scope of Core personnel. However, we can serve as your connection to the Illumina technical support groups for some of these issues if desired. To get some idea of the data analysis issues involved, download this data analysis tech sheet provided by Illumina. An overview of the Genome Studio software and its functionality is offered as part of the genotyping service.
Quality Control
The primary role of the DNA Technologies Core will be to order the materials, run the assays, and carry out initial analyses. A number of sample-independent controls are provided in the assay to monitor different steps in the hybridization and visualization processes, and these are verified to be within specifications for every run. Sample dependent behavior such as call rate and intensity are also examined for appropriate outcomes.
Prices
Prices for running the Infinium assay in our Core are a combination of basic processing costs (labor and non-illumina supplied reagents) plus 5% of the cost of each chip. Because the chips come in various formats, it is not possible to report a common per sample cost, this will vary depending on the format being used. The extra 5% helps cover the cost of replacement chips if needed due to handling or processing issues in the Core.

