Make sure you discuss and plan your experiment with us several weeks in advance and to schedule your cell drop-offs without staff. Contact Hong Qiu for details: honqiu@ucdavis.edu.
We are offering 10X Genomics, SPLit-Seq (ParseBio Whole Transcriptome) & plate-based single-cell sequencing services. Please see the bottom of the page for the latter options.
10X Genomics Single-Cell Omics:
We are working with the latest 10X Genomics chemistries (Next-GEM assays) and the latest equipment (e.g. the temperature-controlled Chromium X processor) to enable:
- Single Cell Gene Expression (3′ GEX V3.1)
- Fixed Single Cell Gene Expression (human and mouse samples)
- Single Cell Immune Profiling (5′ GEX V1.1 and V2 + V(D)J)
- Single Cell ATAC-seq
- HT Gene Expression and Immune Profiling
- Visium Library Preparation (slide preparation not included)
- Single Cell Multiome (combined ATAC + GEX)
A consultation is required prior to scheduling any experiment. We offer free joint consultations with the Bioinformatics core. We can also connect you with your local 10X sales specialists and scientists for office hour appointments!
************************************************************************
UC Davis users with BSL1 samples can access our 10X Chromium controller as part of our shared instruments for customer use. This is advantageous if your group is planning a large number of single-cell experiments. Please contact 10X for training. Due to ongoing COVID19 testing, we have suspended customer instrument use temporarily.
************************************************************************
The 10X Genomics Single Cell suite enables high capture efficiency (of up to 65% of cells loaded) with a flexible workflow, encapsulating 500 to 20,000 cells or nuclei per library together with micro-beads into nano-droplets. (These numbers can be further increased with sample multiplexing.) Each bead is loaded with adapters containing one of 750,000 different barcodes for the single cell RNA-seq library preps. In contrast to other protocols, the 10X controller can load “all” droplets with micro-beads, enabling single-Poisson distribution loading and thus high capture efficiencies (in contrast to double-Poisson loading of other protocols). The single-cell encapsulating process is significantly faster compared to other technologies. Up to sixteen samples can be processed per batch within minutes. The resulting data can be analyzed with the free Cell Ranger and Loupe Cell Browser software. In addition, the Bioinformatics Core has developed a custom single-cell data analysis pipeline for 10X data.
The principles of the 10X Single-Cell RNA-seq library preparation:
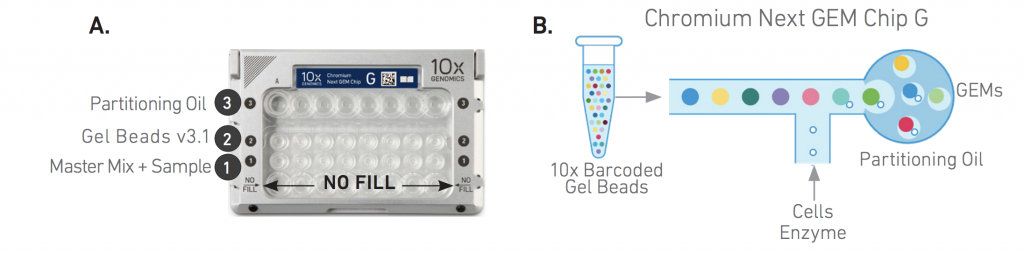
10X Single-Cell Gene Expression (GEX)
10X Chromium Single Cell Features:
- Fast workflow from cell suspension to 3′-cDNA or 5′-cDNA library.
- Captures 100-20,000+ cells per sample and up to 320,000 cells (or ~ 1 million cells with multiplexing) per run in ~20 minutes.
- Recovers up to ~65% of cells (typically 50%).
- Low doublet rate (~0.9% per 1,000 cells).
- Compatible with Illumina and Element Biosciences Aviti sequencers.
The 10X GEX Single-Cell libraries are most economically sequenced on the Illumina NovaSeq 6000 with paired-end reads. (The assay requires at least a 28 cycle forward read, a 10 bp dual index reads, and a 90 cycle reverse read). - For most applications, an average of 40,000 reads per cell should be sequenced (for cell types with complex transcriptomes). For isolated nuclei 25,000 reads each are recommended,
- Dedicated fixed cell assays for human and mouse samples enable scaling up experiments to multiple formaldehyde fixed biological replicates. For other species working with cryo-preserved cells or methanol-fixed cells, enables safe sample shipping and batching.
- 10XGenomics HT assays and chips double both, the number of samples and the number of cells per sample that can be processed simultaneously, compared to the standard assays.
- The cell size limit is comparatively high. Cells can have a diameter of up to 50 µm.
- In addition to cell suspension samples also nuclei suspensions can be studied, enabling the analyses of brain tissues.
- Cell-Plex reagents allow tagging and pooling of human, mouse, and rat cell samples before 10X library prep potentially reducing the costs significantly. This also allows super-loading of a 10X chip channel (with up to 60,000 cells).

Single Cell GEX Resources:
- For information regarding sample preparation, please refer to the 10X cell preparation guide.
- See the 10X Support Center to determine the correct cell numbers and for more information about the workflow.
- For cryopreserved samples please submit at least 100,000 cells per sample.
- 10X has a new demonstrated protocol for methanol fixation, which is compatible with the 10X system.
- The complete User Guides are available here (3′ GEX) and here (5′ GEX).
10X Chromium Single Cell suspension sample requirements:
- Minimum concentration of 100 cells/ul (700 to 1,200 cells/ul optimal range) in a volume of at least 40ul.
- If at all possible, please provide 70ul of single cell suspension (two attempts at chip loading in case of clog plus additional for cell QC). We will require 10 ul sample for the cell counter.
- Maximum concentration of 2000 cells/ul.
- Single-cell suspension should be at least 70% live and free of visible debris and doublets.
- Recommend cell suspension buffer is PBS/0.5% BSA. Cell suspension buffers should be free of EDTA and Mg++ as well as free of DNAse to be compatible with single-cell assay. Up to 2% BSA content is OK. Please see 10X cell preparation guide.
- For fresh samples submitting 40K+ cells total is best.
- For frozen samples, 100,00 cells are the minimum, but 1 million are better.
Our automated cell counters (Luna FL, Logosbio, dual fluorescence, AO/PI) and Countess II (Life Technologies, brightfield, Trypan blue ) can assist you with the preparation and QC of the cell suspension. The cell counters provide total cell counts, report viability, and measure average cell sizes in as little as 10 seconds. Please see the Luna FL guide and the Countess manual.
10X scATAC-Seq
The Chromium Single Cell ATAC Solution allows users to profile the regulatory landscape of chromatin in thousands of cells. Nuclei are transposed in a bulk solution and then are partitioned into nanoliter-scale Gel Beads in-emulsion (GEMs). Please inquire for QC requirements and loading concentration.
- Tested on cell lines, primary cells, fresh, and cryopreserved samples.
- The complete user guide is available here.
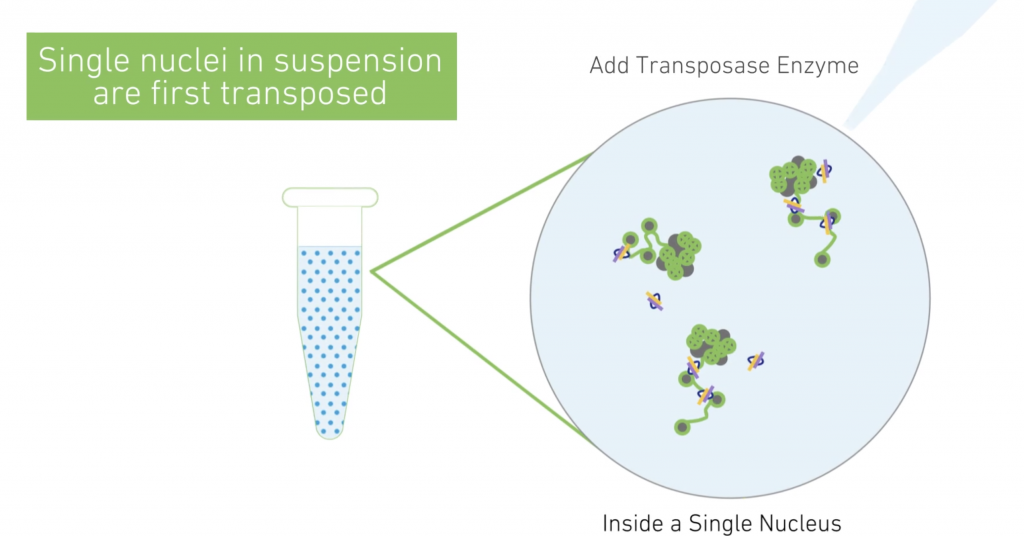
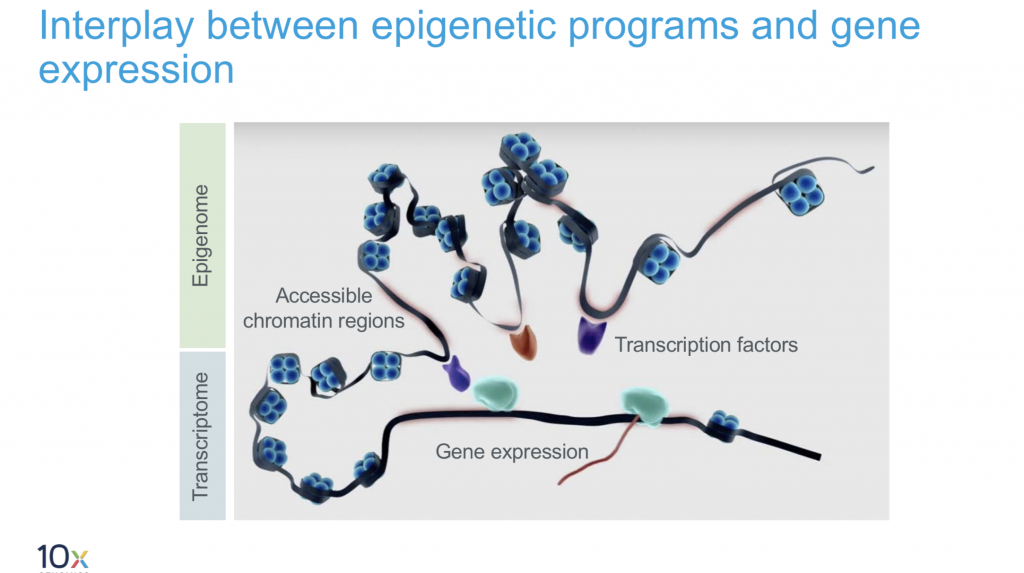
10X Multiome
This is the latest product launch by 10X and promises both gene expression data and regulatory landscape profiling in a single cell.
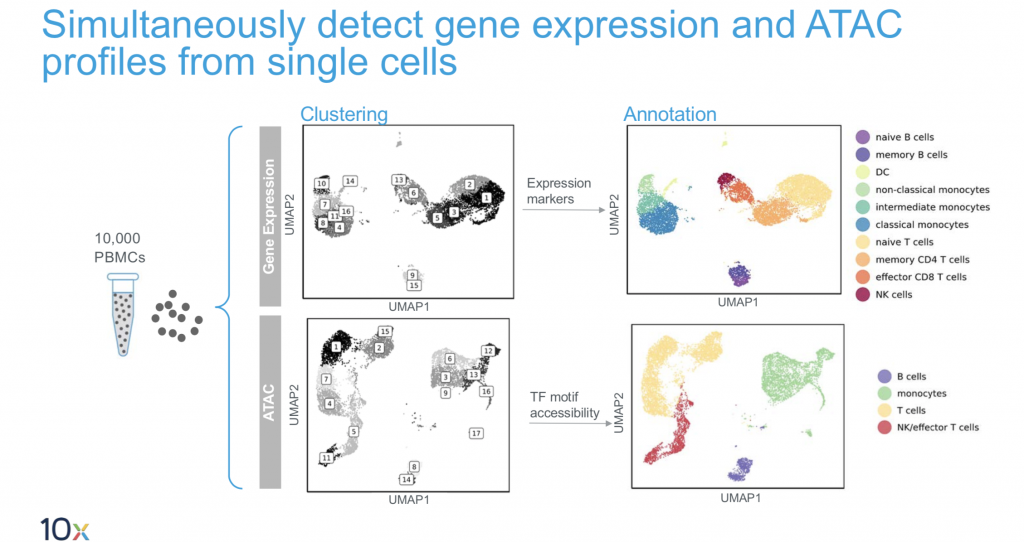
- Input: single nuclei.
- The complete user guide is available here.
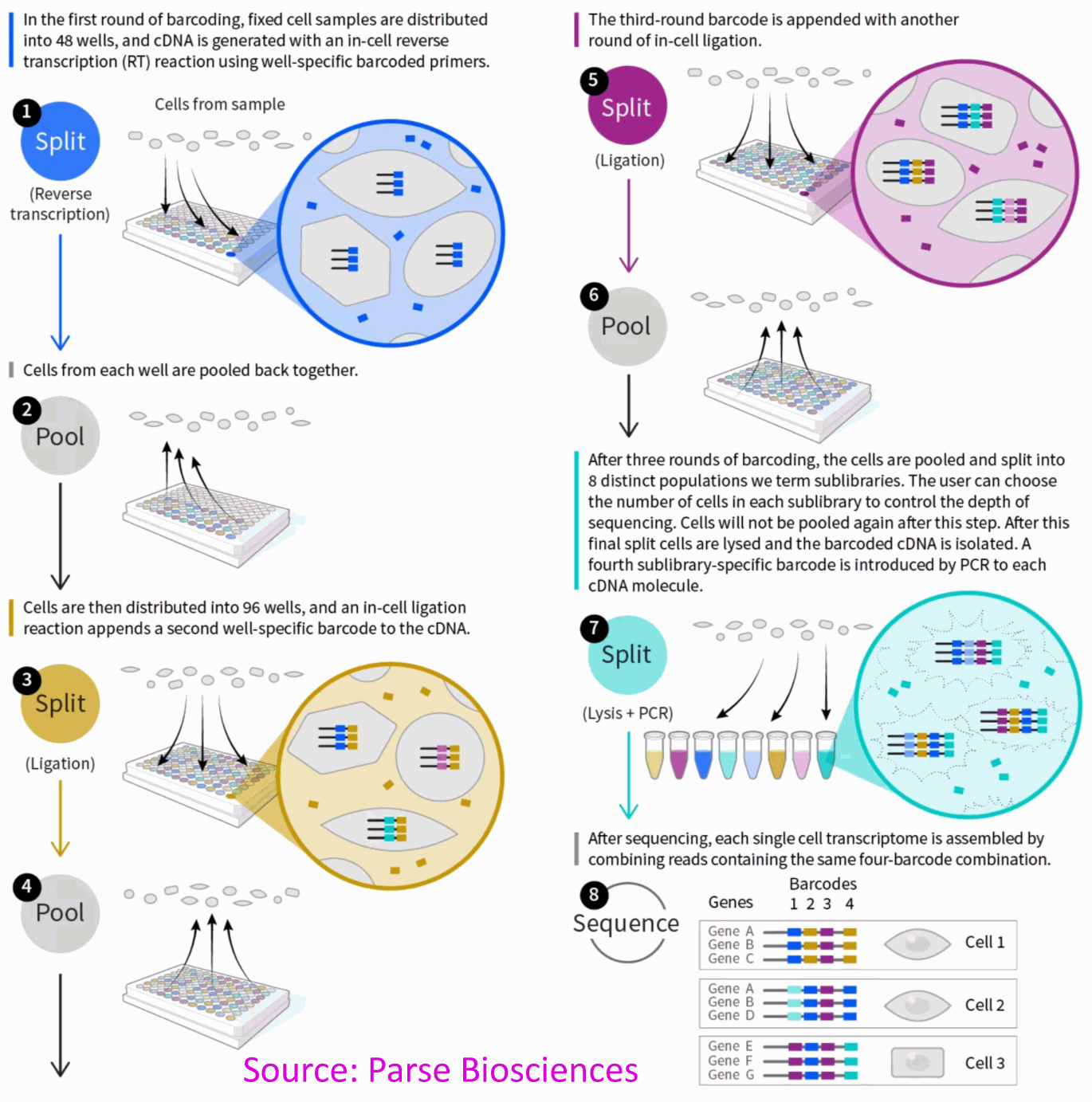
Parse Biosciences Single Cell Whole Transcriptome Solution
The Parse Biosciences Single Cell Whole Transcriptome protocol employs combinatorial indexing of transcripts to identify the cells of origin. It is an improved version of the SPLiT-seq protocol originally developed in the Seelig Lab (Allen Institute for Brain Sciences). At scale it offers lower costs per cell than 10X and requires no specialized equipment; All indexing reactions are performed in 96-well plates. The Parse Bio starting material is fixed samples (single cell or nuclei suspensions), which allows sample storage for months before library preparation as well as safe shipping.