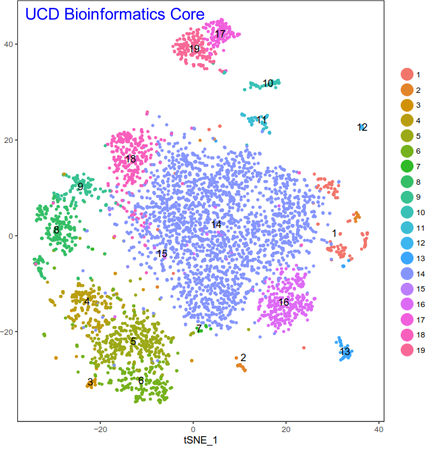
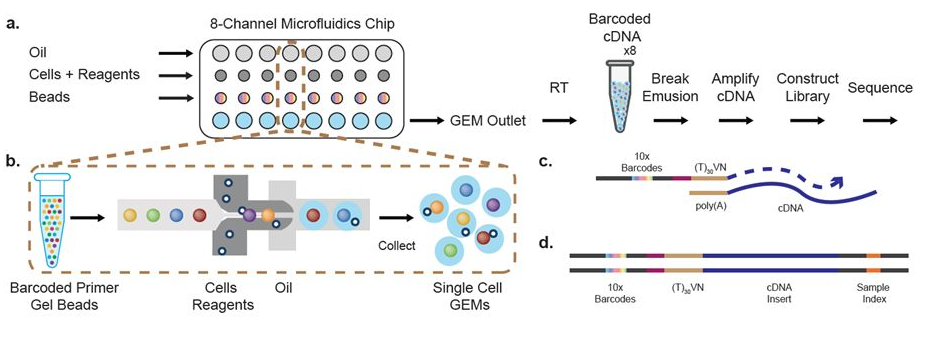
The flexible workflow encapsulates 100 to 15,000 cells or nuclei per library together with micro-beads into nano-droplets. Each bead is loaded with adapters containing one of 750,000 different barcodes for the single cell RNA-seq library preps. In contrast to other protocols, the 10X controller is capable of loading “all” droplets with micro-beads, enabling single-Poisson distribution loading and thus high capture efficiencies (in contrast to double-Poisson loading of other protocols; e.g. Drop-Seq). The resulting data can be analyzed with the free Cell Ranger and Loupe Cell Browser software. In addition the Bioinformatics Core has developed a custom single-cell data analysis pipeline for 10X data.
By default, our staff will prepare the single-cell RNA-seq (scRNA-seq) libraries from cell suspension samples submitted by the research labs. Since the samples should be processed as quickly as possible, the experiments need to be planned and scheduled together with our staff. In case your lab is planning a large number of single-cell experiments, it might make sense for investigators to get trained to be able to process the cell suspensions on the 10X Genomics Chromium controller.
The principles of the 10X Single-Cell RNA-seq library preparation:
10X Chromium Single Cell Features:
- Fast workflow from cell suspension to 3′-cDNA library.
- Captures 100-80,000+ cells in < 10 minutes.
- Recovers up to ~65% of cells.
- Low doublet rate (~0.9% per 1,000 cells).
- Compatible with Illumina HiSeq4000, NextSeq, and MiSeq sequencers.
The 10X Single-Cell libraries are most economically sequenced on the Illumina HiSeq 4000 with paired-end 100bp reads or on the NextSeq with the 150 cycle kits. - For the most common applications 50,000 or more reads per cell should be sequenced.
- It is possible to work with cryo-preserved cells, enabling safe sample shipping and batching.
- In addition to cell suspension samples, nuclei suspensions can be studied.
Please inquire with our 10X Genomics specialist Diana Burkart, PhD, for further details.