 The DNA Tech Core uses two AVITI sequencers for highest quality fast-turnaround short-read sequencing. These sequencers from Element Biosciences provide medium-scale sequencing at higher quality and significantly lower costs compared to the long-time standard Illumina sequencers. While the sequencing chemistry is very different, the instruments are compatible with Illumina libraries and files.
The DNA Tech Core uses two AVITI sequencers for highest quality fast-turnaround short-read sequencing. These sequencers from Element Biosciences provide medium-scale sequencing at higher quality and significantly lower costs compared to the long-time standard Illumina sequencers. While the sequencing chemistry is very different, the instruments are compatible with Illumina libraries and files.
- The AVITI sequencers enable the Core to deliver higher quality sequencing data faster at the similar per-base costs of the biggest flowcell of the much larger NovaSeq 6000.
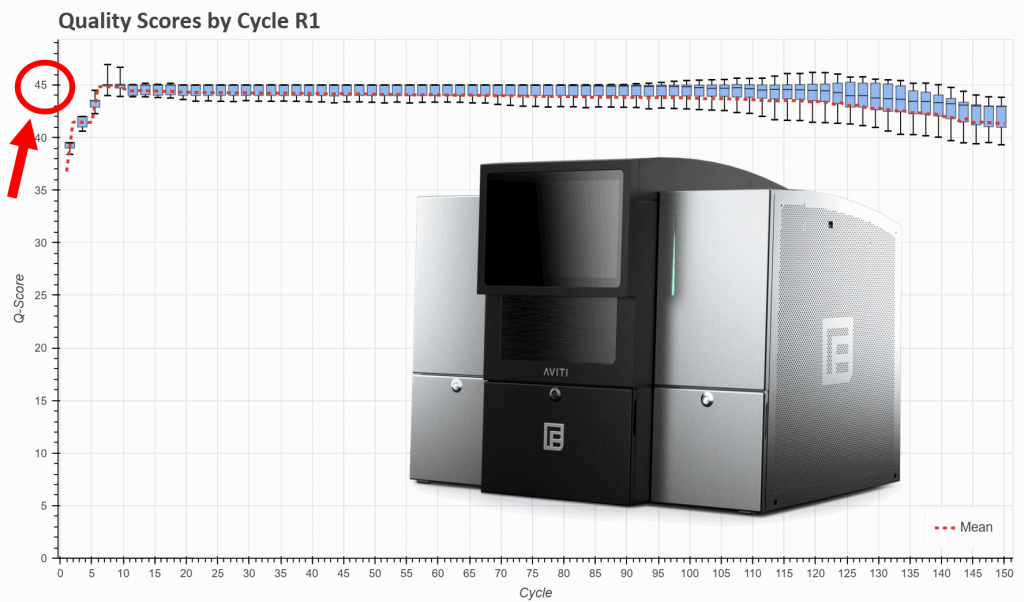
- The AVITI data will have higher quality scores (most bases will have a Phred scale Q-score of 44; equivalent to less than one error in 10,000 bases).
- The AVITI runs two flow-cells independently, with each flow-cell delivering about one billion read pairs.
- Our lab has already more than twelve months of experience with AVITI sequencing.
- The cost for a 300-cycle run (PE150) is $2,000, and $1,400 for a 150-cycle run (UC rates).
- AVITI data will not contain artifacts introduced by the exclusion amplification on patterned flowcells: No index hopping and no read duplication during clustering.
- A 600-cycle kit (enabling PE300 reads) is available and significantly improves on the yields and data quality of the Illumina MiSeq.
Getting started info: > Register with the GC > Sample Requirements > Sample Submission
The main considerations when submitting Illumina libraries for sequencing on the AVITI are:
- the AVITI tolerates longer insert sequencing libraries
- the AVITI requires higher sequencing library concentrations (best submit 20 ul of library pool with a concentration of 16 nM). This is still easy to achieve, especially when pooling several libraries
- the AVITI is compatible with standard Illumina libraries. These libraries will have Truseq-style or Nextera-style adapter sequences as about 99% of the libraries we sequence do. Small RNA-seq libraries will require custom sequencing primers on the AVITI. Not compatible are sequencing libraries with IDT/SwiftBio “Normalase” treatment. Please discuss custom sequencing adapter constructs with us before submitting these
- Avoid submitting libraries with very short inserts — defined as library inserts shorter than the read-lengths of forward or reverse reads.
- Amplify your libraries with a proofreading (high-fidelity) polymerase for your library amplifications (e.g. Kapa HiFi, NEB Q5, Qiagen VeraSeq, …). Libraries will be circularized and A-overhangs created by Taq polymerase would interfere with these.
- We sequence exclusively “Illumina libraries” on the AVITI as they are established in our and most other labs. Element Bio has developed a new proprietary library technology: “Elevate” libraries. Elevate libraries can be generated with kits from the company as well as with Elevate adapters by third parties for many common library prep kits. Please discuss the projects with us before submitting Elevate libraries. We need to know in case you are using Elevate adapters!!!
Unique Sequencing Technology
In contrast to the traditional sequencing-by-synthesis (SBS) chemistry, the AVITI employs a sequencing-by-binding chemistry (SBB) that involves the binding of a multivalent fluorescent polymerase substrate by avidity. The use of these so-called Avidites increases specificity and reduces the fluorescent chemistry costs by an order of magnitude. Due to this approach, the DNA synthesis steps can be carried out with unlabeled nucleotides, further reducing artifacts.
Publications describing the technology and its application:
Arslan et al. 2022: Sequencing by avidity enables high accuracy with low reagent consumption
Biswas et al. 2022: Avidity sequencing of whole genomes from retinal degeneration pedigrees identifies causal variants
Find out more about the AVITI sequencer at: https://www.elementbiosciences.com/products/aviti

