Please note that custom sequencing primers are not supported by Illumina — the company will not replace sequencing kits on run failures. Nevertheless, custom sequencing primers can enable some unique assays. It is certainly worth exploring if assays can’t be converted to use standard Illumina sequencing primers since standard primers are more or less guaranteed to work. Please also see the amplicon sequencing FAQ and note that the client carries the responsibility for any failures due to bad custom primer oligos or poorly designed custom primers.
On MiSeq sequencers custom sequencing primers can be used for three of the reads (forward, reverse, & 1st index reads)
On the NextSeq 550, custom sequencing primers can be used for all four reads (forward, reverse, & 1st and 2nd index reads)
Illumina does not support custom primers on the NovaSeqs at all.
For Illumina Sequencers: Please provide an aliquot for each custom primer (10 ul at 100 uM in EB buffer; low-bind tubes) with each library submission. Custom sequencing primers should be ordered HPLC purified to remove any incomplete oligos.
For the Element Bio AVITI sequencer: Please provide an aliquot for each custom primer (30 ul at 100 uM in low-TE buffer for index 1, index 2 and read 2 sequencing primers; 40 ul at 100 uM in low-TE buffer for read 1 sequencing primers ; low-bind tubes) with each library submission. Custom sequencing primers should be ordered HPLC purified to remove any incomplete oligos.
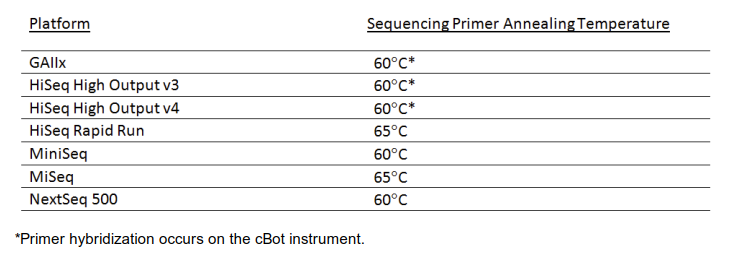
When designing custom sequencing primers, the melting temperature needs to be considered (among other criteria). It is suggested to calculate it with the IDT oligo analyzer for the default buffer conditions (50 mM Na+). Please see the recommended melting temperatures in the image below. While at the oligo analyzer site also check the designs for secondary structures, potential hairpin and self-dimers. In case the melting temperatures of your design are too low, the Tm can potentially be increased by inserting interspersed LNA bases into custom LNA oligos available from Qiagen. Avoid clustering of LNA bases and do not substitute the two last 3′ bases. LNA oligos are significantly more expensive than conventional oligos.
For more info please see this guide from Illumina: miseq-system-custom-primers-guide-15041638-01 and also the index read guide: indexed-sequencing-overview-guide-15057455-04-Illumina-pages1to8
Please note that the 2nd-index read is primed from a flowcell-bound oligo for the Miseq and most other Illumina sequencers. Thus, it cannot be customized. The sequencing of the second index begins with exactly seven dark cycles for which no sequence is recorded. Thus, the first base of the 2nd index sequence has to be the 30th nucleotide from the P5 end of the library molecule for most applications. Please note this FAQ can’t be a comprehensive guide to custom sequencing primer design and usage.
Small RNA-seq / miRNA-seq on the AVITI with libraries employing TruSeq Small RNA sequencing adapters:
Truseq Small RNA libraries will require custom sequencing primers on the AVITI. In most cases there will be no need for a reverse read, thus only TruSeq Small RNA Read 1 and i7/Index 1 custom sequencing primers will be required.
10x Specific recommendations: If using 10X Index Primer Set TS to complete their libraries (i.e. Single Cell Gene Expression Flex or Visium FFPE), they will need to spike-in a TruSeq Small RNA sequencing primer.
| Primer Name | Sequence (5′ -> 3′) | Tm (C) | Length (nt) |
| TruSeq Small RNA Read 1 | ATCTACACGTTCAGAGTTCTACAGTCCGACGATC | 65 | 34 |
| TruSeq Small RNA Read 2 | GTGACTGGAGTTCCTTGGCACCCGAGAATTCCA | 68 | 33 |
| i7/Index 1* | TGGAATTCTCGGGTGCCAAGGAACTCCAGTCAC | 68 | 33 |
| i5/Index 2 | Use Adept Index 2 Primer | — | — |
← FAQ