PacBio Sequel Version 2 Chemistry
We have recently beta-tested the new PacBio Sequel chemistry (V2). The tests have demonstrated significant yield and read-length improvements. All current and future Sequel runs will be carried out with the new chemistry. In addition to the V2 chemistry change, the Sequel software has been updated and allows now for 10 hour movie recordings (previously the limit was 6 hours, as it still is for the RSII). The revised Sequel software also provides usable run-quality metrics.
While read-length metrics are still a bit lower compared to the PacBio RSII, the reduced costs of Sequel sequencing should outweigh the read length disadvantages for the majority of applications, including de novo whole-genome sequencing of bigger genomes.
The example data below are generated from a high quality bird genome sequencing library. The genome is not assembled yet, thus potential artifacts could not be filtered out. Longer read lengths can be achieved on both the RSII and the Sequel when sacrificing yields.
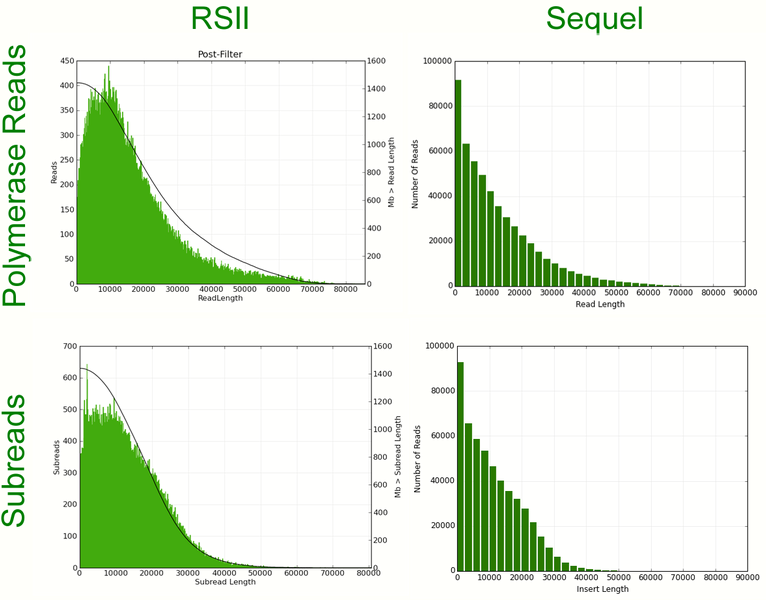
Comparison of run metrics for a bird genome library on RSII and Sequel:
Read length histograms for the same bird genome library on RSII and Sequel:
Titrating the optimal loading concentrations for each library is still more difficult on the Sequel – thus the yields per SMRT-cell still show more variation. After titration, a Sequel SMRT-cell will generate four to five times more data than an RSII SMRT-cell for high quality genomic libraries.
The sequencing of a Sequel SMRT-cell will cost about three times more than that of an RSII SMRT-cell. Since the Sequel requires reduced sequencing library amounts, and thus fewer library preps, sequencing costs will be at least 30% lower per Gb, compared to the RSII for genomes larger than 500 Mb.
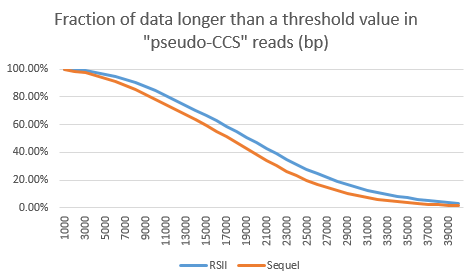
To display the read length differences between the sequencers the “long-subread” read lengths (only the longest subread per ZMW) are plotted below. Specifically, the fraction of data longer than threshold length (x axis) are shown. The RSII shows the biggest advantages if one considers the yield of fragments longer than 22kb. Here the RSII data show an 8% higher content compared to the Sequel. Considering that the cost per Gb of data is likely 30% lower on the Sequel, the Sequel seems to be more economical, even when filtering all reads shorter than 22 kb out of both data sets.
New Iso-Seq Protocol for the Sequel
PacBio will soon provide a new IsoSeq protocol which will make this type of whole-transcript-sequencing analysis a lot more attractive. Due to a reduced bias towards sequencing shorter library molecules on the Sequel, the number of size fractions that should be sequenced separately can be reduced to two (as compared to four or five on the RSII). Similarly, simple IsoSeq experiments without size-selection should generate much more comprehensive data on the Sequel. The new protocol should more than half the costs of typical IsoSeq experiments.
Edit: The Sequel IsoSeq protocol is now available.
Please inquire with us for any Sequel related questions.