NOTE: The Hi-C service is currently suspended.
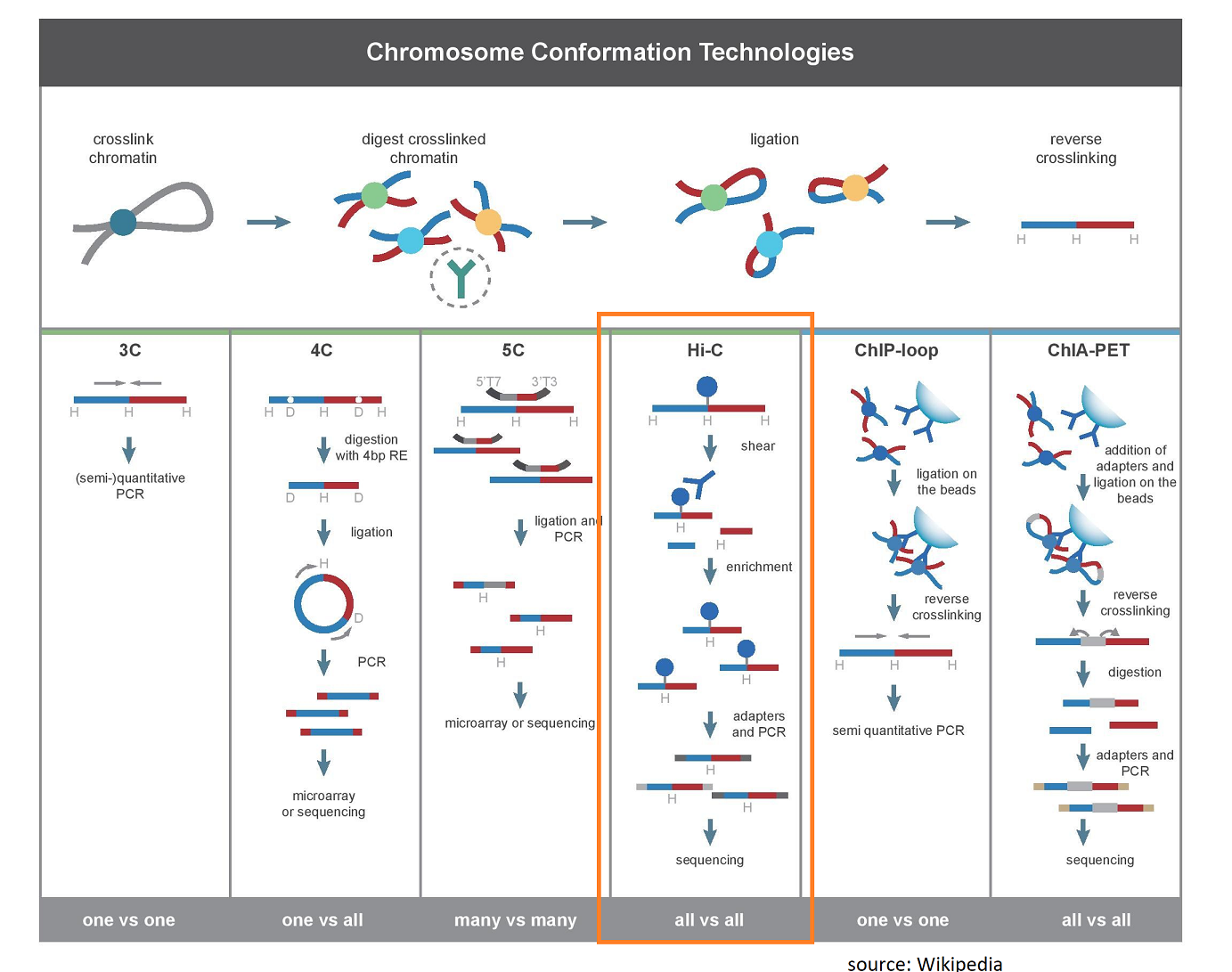
Hi-C is a genome-wide Chromatin Conformation Capture protocol using proximity ligation. The technology is of special interest for two areas of research:
- Three-dimensional genome organization in the nucleus: Please see the excellent review by Andrey & Mundlos “The three-dimensional genome: regulating gene expression during pluripotency and development“. Hi-C allows the analysis of non-random 3d-genome structures, chromatin domains, of which topologically-associating-domains (TADs) are thought to be the most important ones: “TADs have been shown to be a fundamental component of the 3D organization of the genome in the nuclear space, restraining and facilitating enhancer-promoter interactions mostly in a cell type-independent manner” (Andrey & Mundlos, 2017).
- De novo genome assemblies: The two-dimensional genome organization of scaffolds can be derived from the Hi-C 3D chromatin contact data. Thus, Hi-C is the method of choice for the chromosome-scale scaffolding of genome assemblies. Please see, for example, this paper: “Scaffolding of long read assemblies using long range contact information” by Ghurye et al.
The Hi-C libraries are sequenced on Illumina sequencers. At the moment we offer Hi-C for animal samples.
Our Hi-C sequencing expert is Ruta Sahasrabudhe, PhD.
Hi-C Sample Requirements
Cell culture Samples
Please submit 0.5X10^6 cells per experiment. Cells should be pelleted, washed with 1X PBS, supernatant removed, flash frozen in liquid nitrogen and stored at -80C. Please ship the cell pellets on dry ice.
Tissue Samples
Fresh tissue mush be snap frozen in liquid nitrogen right after harvesting and stored at -800C. Frozen sample should be shipped on dry ice. Any freeze thaw cycles should be avoided. Please submit ~50-100mg tissue per experiment. For Hi-C heart, lung and muscles are the preferred tissue types. Liver tissue is not acceptable for Hi-C.