Equipped with the major Next Generation Sequencing (NGS) platforms, the DNA Technologies and Expression Analysis Cores at the UC Davis Genome Center provide comprehensive genome sequencing options for your genome studies. Our portfolio of NGS technologies allows whole genome DNA and RNA sequencing, targeted sequencing (exome, enriched target), and gene regulation (methylation, ChIP, small RNA) studies.
The ability to use NGS technologies as an extension of individual research programs will lead to unprecedented amounts of data and enable novel areas of investigation. Because of the cost, complexity of the process, and pros and cons of each option, careful thought should be given to the experimental design for your specific research goal. We would be happy to discuss this with you.
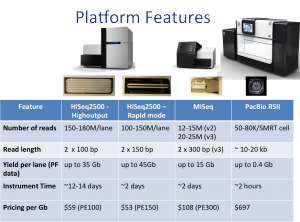
The table below summarizes and compares our NGS platforms and their features (click to enlarge).
Illumina HiSeq 2500
Our Illumina HiSeq2000 was upgraded to HiSeq2500 in June 2013. The HiSeq2500’s rapid mode combines HiSeq data output with MiSeq turnaround time. With this upgrade, we can use either 8-lane flow cell (high-output mode, ~12 days instrument time) or 2-lane flow cell (rapid mode, ~2 days instrument time). High-output mode (using 8-lane flow cell) allows up to PE100. The slow mode provides a greater number of reads and therefore is more appropriate for applications requiring tag counting such as RNAseq. Due to the short instrument time in a rapid mode, read length can be extended to PE250, available only in a rapid mode. Please see table above for performance feature comparison and more details for Illumina sequencing technology here.
Illumina MiSeq
Currently we have three MiSeq to meet the increasing demand. Typical sequencing turnaround time for MiSeq is within 5-7 business days. MiSeq has been popular option for library QC, especially for pool optimization associated with large-scale multiplexing (tag counting and representation), small genome and amplicon sequencing. An exciting feature of the MiSeq is longer read length, currently paired reads of 300 bases (600 cycles, v3 kit), an excellent option for metagenomic sequencing, as well as SR50, PE75 (v3 kit), PE150, and PE250. Illumina is projecting 400 bp PE reads in the near future. Please see table above for performance feature comparison and more details for Illumina sequencing technology here.
PacBio RS II
PacBio RS II (Pacific Bioscience Real-time Sequencer), a single-molecule long-read sequencer, has been used for sequencing for bacteria, BACs, plasmids, viruses, phages, and amplicons. PacBio is the only technology available today that provides transcript read lengths from a single molecule (without PCR amplification). This allows detection of gene fusions and rearrangement events, and read-through of genomic regions with low complexity such as long repeats and regions of high GC content. PacBio is an excellent option for providing scaffolds for de novo assembly as well. Detection of kinetic changes (methylation) can be carried out using current chemistry and software. Recent improvement with magnetic bead loading (preferential loading of longer fragment), several rounds of chemistry upgrades (C1 to C2 to P4), and imaging of entire 150k ZMW (zero-mode waveguides where sequencing is happening) provides an average median read length of ~5 kb with long reads tailing off to 10kb, up to 20kb. Accuracy is currently ~88% and errors are primarily associated with insertion but otherwise random (no systematic bias). The current rate of 12% errors over 1 kb is considered equivalent to a mappable length of greater than 750 bp. The longer reads also allow CCS (Circular Consensus Sequencing) to provide highly accurate long reads. Their latest P5-C3 chemistry promises average reads of over 8 kb and up to 30 kb. Please see table above for performance feature comparison to Illumina sequencing platform and more details for PacBio RSII sequencing technology here.

